Multi-task dynamical systems: learning a family of sequences
The multi-task dynamical system (MTDS) was the focus of my phd. In this post, the first of a two-part series, I’ll try to unpack the motivation for the project, as well as providing a concise description of the model.
Sequence families
Physical models are ubiquitous in (so-called ‘hard’) scientific disciplines. At a macro level, nature appears to obey some remarkably simple rules, which can be exploited to provide forecasts of physical phenomena with high accuracy. These rules are codified into models with parameters which ‘tune’ the model to a given situation, such as lengths, masses or damping factors. Under all the different parameter configurations, such a model corresponds to a collection of sequences which we will call the ‘sequence family’. Examples include bouncing balls (with the family corresponding to different gravitational fields or drag coefficients); or damped harmonic oscillation (under different frequencies and/or decay coefficients) – see Figures 1a, 1b below.


Sequence families also exist in many real-world situations where the data generating process is poorly understood. This case is evidently the norm, not the exception, and is apparent in such domains as healthcare, finance, retail, and graphics (e.g. mocap). For instance, the person-to-person differences in ECG waveforms and store-to-store differences in retail sales (Figures 2a and 2b) are – to the best of my knowledge – not well understood. Nevertheless, we are often capable of curating many examples to represent the sequence family.


Where a model of the sequence family is available, forecasting (or otherwise modelling) a sequence can be tailored to the individual. This is useful for modelling the sales response of a product, or more crucially, modelling the response of a patient to a drug. However, where no sequence family model is available, such tailored predictions are not so easily available.
The inductive bias of a sequence family
Let’s suppose one wanted to predict the trajectory of a bouncing ball from a small number of observations, and further, suppose that the material properties of the ball were not known. Figure 3 shows three observations of the height of a ball denoted by black crosses. If we assume some measurement error, there are infinitely many sequences from the ‘bouncing ball’ family which can fit the data, some examples of which are drawn below.
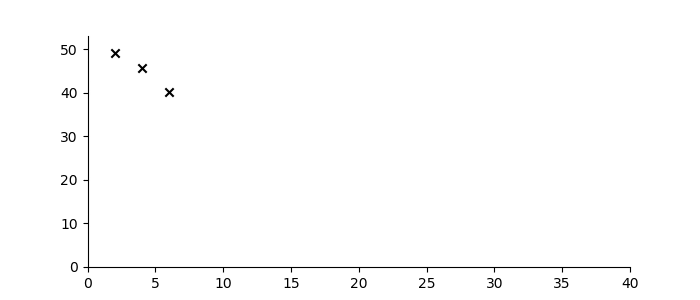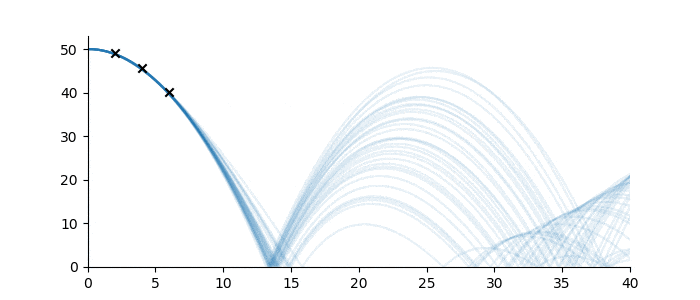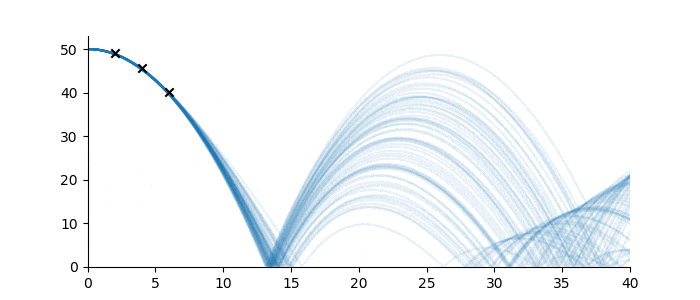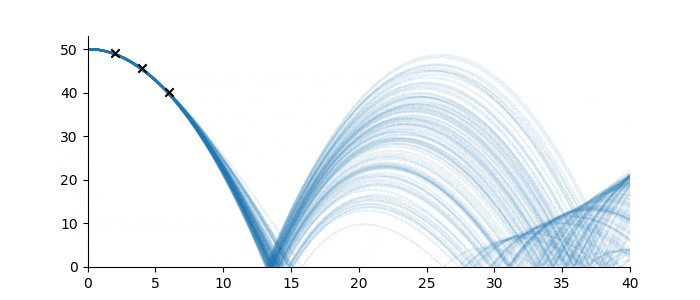
Figure 3: Bouncing ball sequence family members (blue) which are consistent with the three observations (black).
Despite the infinitude of possible sequence completions, we still have a good idea of how the ball will move in the short term, and a qualitative idea of its continued motion. This follows from the strong inductive bias imposed by the sequence family.
Suppose, in contrast, that no sequence family was known for these observations, and hence no inductive bias was available. What then could be said? In this case, almost nothing at all; the sequence continuation might be just about anything. Even just to visualize the problem, we must impose a weak inductive bias, such as once-differentiable sequences with a ‘sensible’ scale length and magnitude. See below for some examples.1
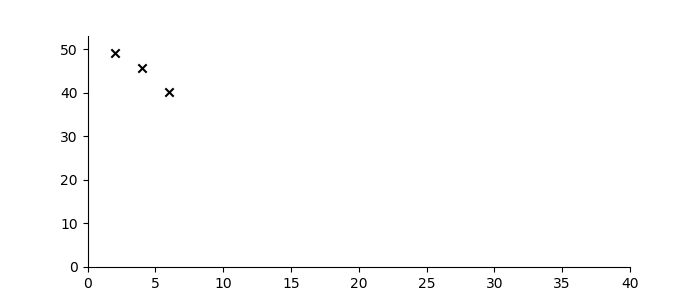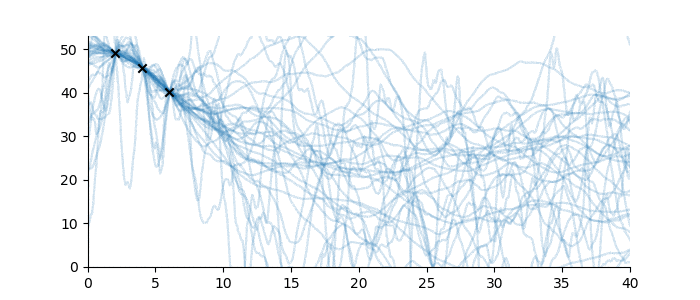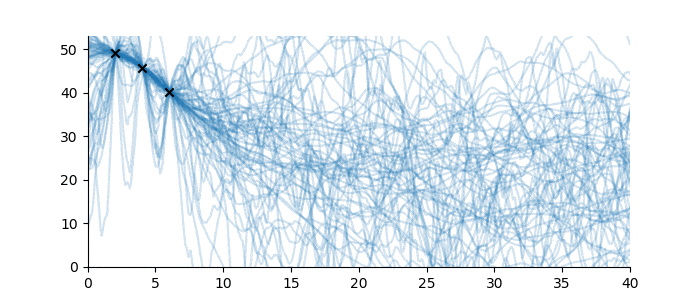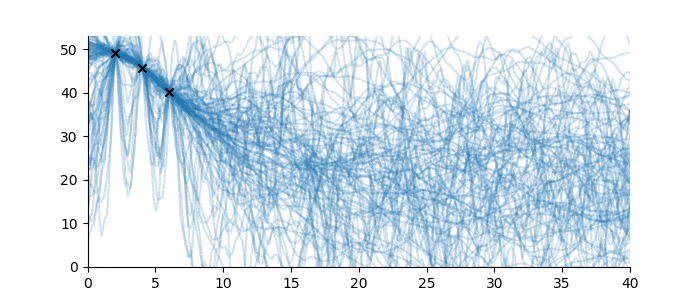
Figure 4: Sequences which are consistent with the three observations without a known sequence family.
We may as well give up on forecasting in this case. Access to an inductive bias gives us the answers to the following crucial questions:
- Is the past indicative of the future?
- If so, in what way(s)?
- Can these be sufficiently well determined, given the data, to make meaningful predictions?
In the absence of an inductive bias, even the first question cannot be answered affirmatively. Ultimately, forecasting sequential data is only possible via use of an inductive bias about the data generating process, and only useful if this inductive bias is well-matched to the true process.2 In the case of the physical models discussed, the inductive bias encodes a good approximation of the true process, allowing excellent predictions.
Modelling the sequence family
What does a model of the sequence family look like? A sequence family can be modelled hierarchically, with each member of the family allocated its own sequence model. This corresponds to a likelihood $p(\mathbf{y}_{1:T}\,|\, \boldsymbol{\theta})$ (ignoring inputs for the time being), with a different parameter $\boldsymbol{\theta}^{(i)} \in \Theta$ for each sequence $i$. In the case of the bouncing ball, the likelihood is the solution to a differential equation ($\boldsymbol{\theta}^{(i)}$ corresponding to specific values of gravitational force and drag), plus some Gaussian noise. These individual parameters $\boldsymbol{\theta}^{(i)}$ are themselves modelled with a prior distribution $p(\boldsymbol{\theta})$. This defines the variability between sequences in the sequence family: a tightly concentrated prior will result in little inter-sequence variation, and an uninformative prior may result in large differences between members of the family. The hierarchical model of the sequence family can be expressed as:
For meaningful sequence families, the typical set of $p(\mathbf{y}_{1:T})$ is small compared to the possible sequence space $\mathbb{R}^{n_y \times T}$: the strength of the inductive bias depends directly on the relative size of the typical set. For the physical models, scientific investigation has provided us with the likelihood $p(\mathbf{y}_{1:T}\,|\, \boldsymbol{\theta})$, and the prior may be specified using domain knowledge since the parameters are directly interpretable. In the more general setting, neither of these quantities are easily specified. Let us first consider the likelihood: I propose using a dynamical system; a general purpose choice.
Dynamical systems
Let’s briefly review what a dynamical system is.34 Dynamical systems, or state space models posit a latent (unobserved) chain of random variables $\mathbf{x}_t$, $t=1,\ldots,T$ which account for the time-structured evolution of the sequence. Crucially, the observed $\{\mathbf{y}_t\}$ only give a partial insight into this evolution, allowing the state $\{\mathbf{x}_t\}$ to capture the relevant information from the past. The state $\mathbf{x}_t$ can often function as a bottleneck, removing irrelevant historical information, allowing a parsionious representation of the sequence $\mathbf{y}_{1:t}$ to date.

Figure 5: graphical model of a dynamical system.
Dynamical systems take the following form, where the hidden state $\mathbf{x}_t$ follows a (possibly stochastic) dynamical model, and the $\mathbf{y}_t$ are conditionally independent of the past, given $\mathbf{x}_t$:
for $t=1,\ldots, T$, with parameters $\boldsymbol{\theta} = \{ \boldsymbol{\psi},\, \mathbf{x}_0\}$. The distribution over the observations can then be obtained via marginalization (integration):
Dynamical systems have a number of desirable properties in general, such as time-invariant feature extraction, linear complexity in $T$, and in principle, an unbounded length of temporal dependence. The class of dynamical systems is highly general and encompasses ARMA type models, linear Gaussian state space models, some GPs, recurrent neural networks (RNNs) and others beside. Dynamical systems may often also contain inputs ($\mathbf{u}_t$), as in the graphical model shown in Figure 5.
Learning a model of the sequence family
There is a certain degeneracy between the likelihood and the prior. One may consider a highly over-parameterized likelihood, and set all the unnecessary parameters to zero in the prior. This is a useful approach, since it largely avoids the search over architecture choice, and results in a single learning problem. As such, let us consider a large dynamical system; with the right parameters, such models can do a good job of modelling many real-world phenomena. However, the corollary of this is the presence of a weak inductive bias, which puts us back into the high-variance prediction situation of Figure 4.
In order to circumvent this problem, a common approach is to pool together the collected sequences, and a single model with $\boldsymbol{\theta}=\boldsymbol{\theta}_0$ used to fit them all. This is effectively appealing to some form of averaging to reduce the high-variance predictions. We call this one-size-fits-all approach a ‘pooled model’, and this corresponds to using a prior $p(\boldsymbol{\theta}) = \delta(\boldsymbol{\theta} - \boldsymbol{\theta}_0)$; a Dirac delta function.5 This degenerate sequence family is clearly unable to model the complexity of the inter-sequence variation and can dramatically underfit the sequences. For instance, personalized predictions are not possible. I must stress that this is a very common approach.
Recurrent neural networks (RNNs) are widely trained as a ‘pooled model’, and nevertheless can model the inter-sequence variation well (if not entirely reliably). But this performance does not generally extend to linear dynamical systems, ARMA models, or any other commonly used statistical or engineering models. In consequence of using an RNN, we need large amounts of data, and lose hope of interpreting the model, or obtaining the kinds of insight provided by structured and switching dynamical systems. Further discussion of RNNs and our related contributions will be relegated to the next post; for now we will consider the common case where interpretation, unsupervised insight and low sample complexity are important factors.
One may instead take a scientific approach, and through careful examination and experimentation derive a bespoke model $p(\mathbf{y}_{1:T}\,|\, \boldsymbol{\theta})$ for the application, with a small number of interpretable parameters. But in many cases this will be impractical, in terms of time or money, and sometimes perhaps impossible.
Instead of these standard approaches, we ask: can we learn the entire hierarchical model – the sequence family, rather than just a single model? The multi-task dynamical system (MTDS) was developed to answer this question. Instead of learning a single parameter $\boldsymbol{\theta}_0$ via averaging, the MTDS learns a distribution $p(\boldsymbol{\theta})$. This distribution is assumed to have support only on a low dimensional manifold since we will rarely have the statistical strength to learn a distribution over all of $\Theta$. This captures the important degrees of freedom in the sequence family, and ignores the others. Hence we learn to approximate the inductive bias implied by the training set, and prune out inter-sequence variation which is not supported by the training set. While not our motivation, this can also remove problematic ‘sloppy directions’ (see e.g. Transtrum et al., 2011); directions in parameter space which make little difference to the model fit.

The multi-task dynamical system (MTDS)
Having motivated the MTDS, we now come to describe it mathematically. As we have seen, the MTDS is a hierarchical model of dynamical systems. We will define the parameter prior by:
which restricts $\boldsymbol{\theta}$ to a manifold defined by $h_{\boldsymbol{\phi}}$, indexed by the latent variable $\mathbf{z} \sim p(\mathbf{z})$. The (possibly nonlinear) mapping $h_{\boldsymbol{\phi}}$ embeds the latent variable in $\Theta$, and defines a low dimensional manifold when $\textrm{dim}(\mathbf{z}) < d$. Each sequence $i$ in the training set draws a parameter vector $\boldsymbol{\theta}^{(i)} \sim p(\boldsymbol{\theta}; \boldsymbol{\phi})$, with an associated latent variable $\mathbf{z}^{(i)}$. Hence the generative model for each sequence $i \in 1,\ldots, N$ is:
for $t = 1,\ldots, T_i$. Our experiments have used $p(\mathbf{z}) = \textrm{Normal}(\mathbf{z}\,\vert\, \mathbf{0}_k, I_k)$, which allows a factor analysis or VAE6-like prior over $\boldsymbol{\theta}$ depending on the choice of $h_{\boldsymbol{\phi}}$. The initial state $\mathbf{x}_0$, may be learned, fixed to some value (e.g. to $\mathbf{0}$) or made dependent on $\mathbf{z}$.

Figure 7: dynamical system approaches: (left) single task; (middle) multi-task; (right) pooled.
This hierarchical construction sits between the two common extremes of time series modelling: either
learning separate models per sequence (Figure 7, left) or pooling all the sequences and learning a single
model (Figure 7, right). The MTDS learns a manifold (described by $h_{\boldsymbol{\phi}}$) in parameter
space, with the goal of capturing a small number of degrees of freedom of the sequence model. By maximizing
the marginal (log) likelihood:
the MTDS can learn a distribution $p(\boldsymbol{\theta}; \boldsymbol{\phi})$ which is hopefully a good approximation of the true sequence family. Details about optimizing eq. (\ref{marginalllh}) can be found on arxiv (and soon in my thesis); for certain base dynamical systems, it suffices to use variational methods, but some circumstances require a little more care.
Example: Linear dynamical systems
Nice in theory. Let’s take it for a ride. As an example, consider the MTDS constructed from a linear dynamical system (LDS) with a state dimension of $n_x=8$, inputs $\mathbf{u}_t \in \mathbb{R}^{n_u}$, and emissions $y_t \in \mathbb{R}$ for each $t \in 1,\ldots,T$. For the sake of ease, we will assume the LDS has a deterministic state. The LDS is defined as:
for $\epsilon_t \sim \mathcal{N}(0, \sigma^2)$. Such a model has parameters $\boldsymbol{\theta} = \{A, B, C, D, \sigma\}$, with $\boldsymbol{\theta} \in \mathbb{R}^d$ where $d= 74 + 9n_u$. While this is small by deep learning standards, it’s relatively large compared to many physical models.
Suppose, for example, that we are only interested in sequences described by the linear combination of two damped harmonic oscillators, with no dependence on inputs. Using a weak prior distribution over $\boldsymbol{\theta}$, e.g. $p(\boldsymbol{\theta}) = \mathcal{N}(\mathbf{0}_d, \kappa I_d)$ for $\kappa \in \mathcal{O}(10)$ results in a fairly low inductive bias; such a model can describe a wide variety of possible sequences (see samples in Figure 8a)7. In contrast, learning a $p(\boldsymbol{\theta}; \boldsymbol{\phi})$ under the framework of the MTDS on a 4-dimensional manifold results in a strong inductive bias (see samples in Figure 8b).


The learned model is an approximation of the true sequence family, and can be used to fit novel sequences with relatively small amounts of data. In principle, the MTDS can learn a model that performs as well as a true physical model, but importantly, can yield similar gains even where no physical model is known (provided the signal-to-noise ratio is similarly high). In my thesis, we demonstrate that we can learn physical models well, via use of the damped harmonic oscillation example introduced above. To get a feel for how these models can be useful in practice, see below for a video of how quickly the MT-LDS can tune into novel sequences. The iterative updates are performed via Bayesian updating, with 95% credible intervals shown in orange.
For more details on how the LDS is parameterized, and how the Bayesian inference is implemented, see the paper on arxiv for the time being – I’ve swept a few details under the rug. I’m currently writing this up in a more complete form in my thesis.
Implementation
This notebook provides an extensive exploration of the performance of the MT-LDS, including the 8-dimensional example above. This builds on the implementation at https://github.com/ornithos/MTDS.jl which is a work-in-progress to consolidate the codebase for my phd. The sequential inference routine has been factored out, and is now available as a standalone inference library.
Bibliography
- Särkkä, S. (2013). Bayesian Filtering and Smoothing (Vol. 3). Cambridge University Press.
- Kingma, D. P., & Welling, M. (2014). Stochastic Gradient VB and the Variational Auto-Encoder. Second International Conference on Learning Representations, ICLR.
- Transtrum, M. K., Machta, B. B., & Sethna, J. P. (2011). The Geometry of Nonlinear Least Squares with Applications to Sloppy Models and Optimization. Physical Review E, 83(3).
Footnotes
-
Full disclosure: the sequences drawn in Figure 4 use a Gaussian Process with Matérn 3/2 covariance function, with magnitude 15 and various scale lengths. For the purposes of the illustration, this uses a constant mean function at height 20. This is technically a sequence family too, but a very large one. None of this is pertinent to the discussion. ↩
-
Of course, this cannot be known in advance except in a few special cases, and hence is a matter of trust. It has long been noted that predicting sequential data (or ‘forecasting’) is a problematic pursuit. Of the many famous quotations on the subject (see e.g. the Exeter forecasting quotes page), one that I particularly like is from Peter Drucker: “[Forecasting] is like trying to drive down a country road at night with no lights while looking out the back window.” ↩
-
This is not a great introduction for those who are unfamiliar with them – I highly recommend Särkkä (2013) as an introductory text, although it is not necessary to fully understand dynamical systems for what follows. It is sufficient to understand that they are flexible sequential models. ↩
-
In this article, I use the term dynamical systems primarily to refer to their discrete time formulation. This simplifies some of the explanation and mathematical machinery, but the discussion applies more widely. ↩
-
Pooled models may be a little more complicated, for instance via use of time warping or random effects governing the signal magnitude. But in the vast majority of cases, the shape of the modelled sequence is only an average. ↩
-
VAE = Variational autoencoder (Kingma & Welling, 2014). ↩
-
While Figure 8a looks a little like white noise, it is in fact a weak prior on the specified MT-LDS. The eye tends to be drawn to the high frequency components, which are dominant with high probability under the chosen prior. ↩
